

David Barford
Mechanism and regulation of chromosome segregation in mitosis
dbarford@mrc-lmb.cam.ac.uk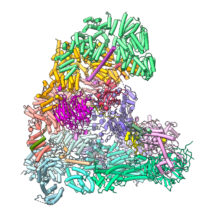
Our research is aimed at understanding the mechanisms and regulation of chromosome segregation in mitosis. During the cell cycle, accurate chromosome segregation ensures the correct inheritance of genetic material and organismal survival. Errors in this process cause aneuploidy leading to cancer and developmental defects. Duplicated sister chromatids are segregated in mitosis by a large cellular apparatus, the bipolar mitotic spindle. At metaphase, condensed chromosomes are aligned on the metaphase plate at the centre of the mitotic spindle. Each sister chromatid of the chromosome is attached to microtubules by kinetochores, large protein complexes that specifically assemble at the centromere. Once all chromosomes attach to the spindle, and tension is exerted at the kinetochore-microtubule attachment site, anaphase is triggered. This results in the loss of sister chromatid cohesion and the segregation of each sister chromatid to opposite poles of the cell, a process powered by microtubule depolymerization. Anaphase onset is triggered by the anaphase-promoting complex (APC/C), a large multi-subunit complex (~1.2 MDa) that functions as an E3 ubiquitin ligase to regulate different stages of the cell cycle. In earlier work we determined cryo-EM structures of the APC/C, explaining how the APC/C recognizes and ubiquitinates its substrates, and how its activity is controlled by protein phosphorylation and the spindle assembly checkpoint
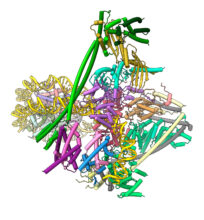
Our current focus is to understand the process of chromosome segregation mediated by the kinetochore. We are undertaking a programme to determine the kinetochore’s molecular and cellular structure and mechanism. We use a variety of approaches, including complete reconstitution of kinetochores and their attachment to chromosomes and microtubules for analysis by cryo-electron microscopy and biophysical methods such as optical tweezers. We are also investigating the structure of the kinetochore in situ using cryo-electron tomography. We recently determined cryo-EM structures of the inner kinetochore (CCAN) bound to centromeric CENP-A nucleosomes. In the future, we aim to determine cryo-EM structures of complete reconstituted human and budding yeast kinetochore complexes that will aid their visualization at centromeres in cells. We also seek to understand the spindle assembly checkpoint and error correction checkpoint from a structural, biochemical and biophysical mechanistic perspective.
Potential PhD students and post-docs interested in working on these questions are encouraged to contact David Barford directly.
Human inner kinetochore – DNA
Selected Papers
- Muir, K.W., Batters, C., Dendooven, T., Yang, J., Zhang, Z., Burt, A., Barford, D. (2023)
Structural mechanism of outer kinetochore Dam1-Ndc80 complex assembly on microtubules
Science 382: 1184-1190 - Dendooven, T., Zhang, Z., Yang, J., McLaughlin, S.H., Schwab, J., Scheres, S.H.W., Yatskevich, S., Barford, D. (2023)
Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere
Sci. Adv.eadg7480. doi: 10.1126/sciadv.adg7480. - Yatskevich, S., Muir, K.W., Bellini, D., Zhang, Z., Yang, J., Tischer, T., Predin, M., Dendooven, T., McLaughlin, S.H., & Barford, D. (2022)
Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science 376: 844-852. - Yan, K., Yang, J., Zhang, Z., McLaughlin, S.H., Chang, L., Fasci, D., Ehrenhofer-Murray, A.E., Heck, A.J.R. and Barford, D. (2019)
Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature 574: 278-282 - Yan, K., Yang, J., Zhang, Z., McLaughlin, S.H. and Barford, D. (2018)
Architecture of the kinetochore CBF3-CEN3 DNA complex of budding yeast.
Nature Structural Molecular Biology 25: 1103-1110 - Alfieri, C., Chang, L. and Barford, D. (2018)
Structural basis for the remodelling of the cell cycle checkpoint protein Mad2 by the AAA+ ATPase TRIP13.
Nature 559: 274-278. - Alfieri, C., Zhang, S. and Barford, D. (2017)
Visualizing the complex functions and mechanisms of the anaphase-promoting complex/cyclosome (APC/C).
Open Biol. (review).: pii: 170204. doi: 10.1098/rsob.170204 - Alfieri, C., Chang, L., Zhang, Z., Yang, Y., Maslen, S., Skehel, M. and Barford, D. (2016)
Molecular basis of APC/C regulation by the spindle assembly checkpoint.
Nature 536: 431-436 - Zhang, S., Chang, L., Alfieri, C., Zhang, Z., Yang, J., Maslen, S., Skehel, M. and Barford, D. (2016)
Molecular mechanism of APC/C activation by mitotic phosphorylation.
Nature 533: 260-264 - Chang, L., Zhang, Z., Yang, J., McLaughlin, S.H. and Barford, D. (2015)
Atomic structure of the APC/C and its mechanism of protein ubiquitination.
Nature 522: 450-454
Group Members
- Dom Bellini
- Forson Gao
- Paula Paredes Vergara
- Liting Wei
- Yvonne Winterborn
- Jing Yang
- Cong Yu
- Ziguo Zhang